Summary stats with dplyr
- get DWD data
- read data
- convert data
- plot some data
- calculate mean for month
- calculate mean for yday
- precipitation
Here I show how to do some simple summary statistics with the dplyr package.
We will be using the followint packages:
library(lubridate)
library(dplyr)
library(readr)
library(ggplot2)get DWD data
Files can be downloaded and unzipped by hand from https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical. Or you can use the following script:
data_dir = "data/dwd"
dir.create(data_dir)
urls <- c(Cottbus="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_00880_18870101_20231231_hist.zip",
Warnemünde="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_04271_19470101_20231231_hist.zip",
#Freiberg="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_01441_19450701_19930430_hist.zip",
#Stuttgart="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_04927_19490801_19840731_hist.zip",
"Garmisch-Patenkirchen"="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_01550_19360101_20231231_hist.zip",
Zugspitze="https://opendata.dwd.de/climate_environment/CDC/observations_germany/climate/daily/kl/historical/tageswerte_KL_05792_19000801_20231231_hist.zip")
for(url in urls){
destfile = file.path(data_dir, basename(url))
if(!file.exists(destfile)){
download.file(url, destfile)
unzip(destfile, exdir = data_dir)
}
}read data
We use the readr package to read the csv files:
data_files <- list.files(data_dir, pattern="produkt_klima_tag", full.names = T)
data <- read_delim(data_files, delim = ";", na = "-999", trim_ws = T) We use list.files to list all files starting with “produkt_klima_tag” in the data directory. The read_delim function reads all files into a single R object. Stations can be identified by their STATIONS_ID in the dataset.
See what’s in the dataset:
data## # A tibble: 153,294 × 19
## STATIONS_ID MESS_DATUM QN_3 FX FM QN_4 RSK RSKF SDK SHK_TAG
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 880 18870101 NA NA NA 1 0 0 NA 0
## 2 880 18870102 NA NA NA 1 0 0 NA 0
## 3 880 18870103 NA NA NA 1 0 0 NA 0
## 4 880 18870104 NA NA NA 1 0 0 NA 0
## 5 880 18870105 NA NA NA 1 0 0 NA 0
## 6 880 18870106 NA NA NA 1 0 0 NA 0
## 7 880 18870107 NA NA NA 1 3.3 4 NA 4
## 8 880 18870108 NA NA NA 1 0 0 NA 4
## 9 880 18870109 NA NA NA 1 0 0 NA 4
## 10 880 18870110 NA NA NA 1 0 0 NA 4
## # ℹ 153,284 more rows
## # ℹ 9 more variables: NM <dbl>, VPM <dbl>, PM <dbl>, TMK <dbl>, UPM <dbl>,
## # TXK <dbl>, TNK <dbl>, TGK <dbl>, eor <chr>convert data
Convert date:
data <- data %>%
mutate(MESS_DATUM = as.POSIXct(as.character(MESS_DATUM), format="%Y%m%d")) %>%
mutate(STATIONS_ID = as.factor(STATIONS_ID))The %>% notation and mutate function comes from the dplyr package.
Calculate yday and month:
data <- data %>%
mutate(yday = yday(MESS_DATUM)) %>%
mutate(month = month(MESS_DATUM))The yday and month functions come from the lubridate package
Add station names:
stations <- tribble(
~STATION_NAME, ~STATIONS_ID,
"Cottbus", "880",
"Warnemünde", "4271",
"Garmisch-Patenkirchen", "1550",
"Zugspitze", "5792"
)
data <- data %>% left_join(stations)plot some data
We plot data with the ggplot2 package:
data %>%
ggplot() +
aes(x=yday, y=TMK) +
geom_point(size=0.1, alpha=0.1) +
facet_wrap(STATION_NAME~.)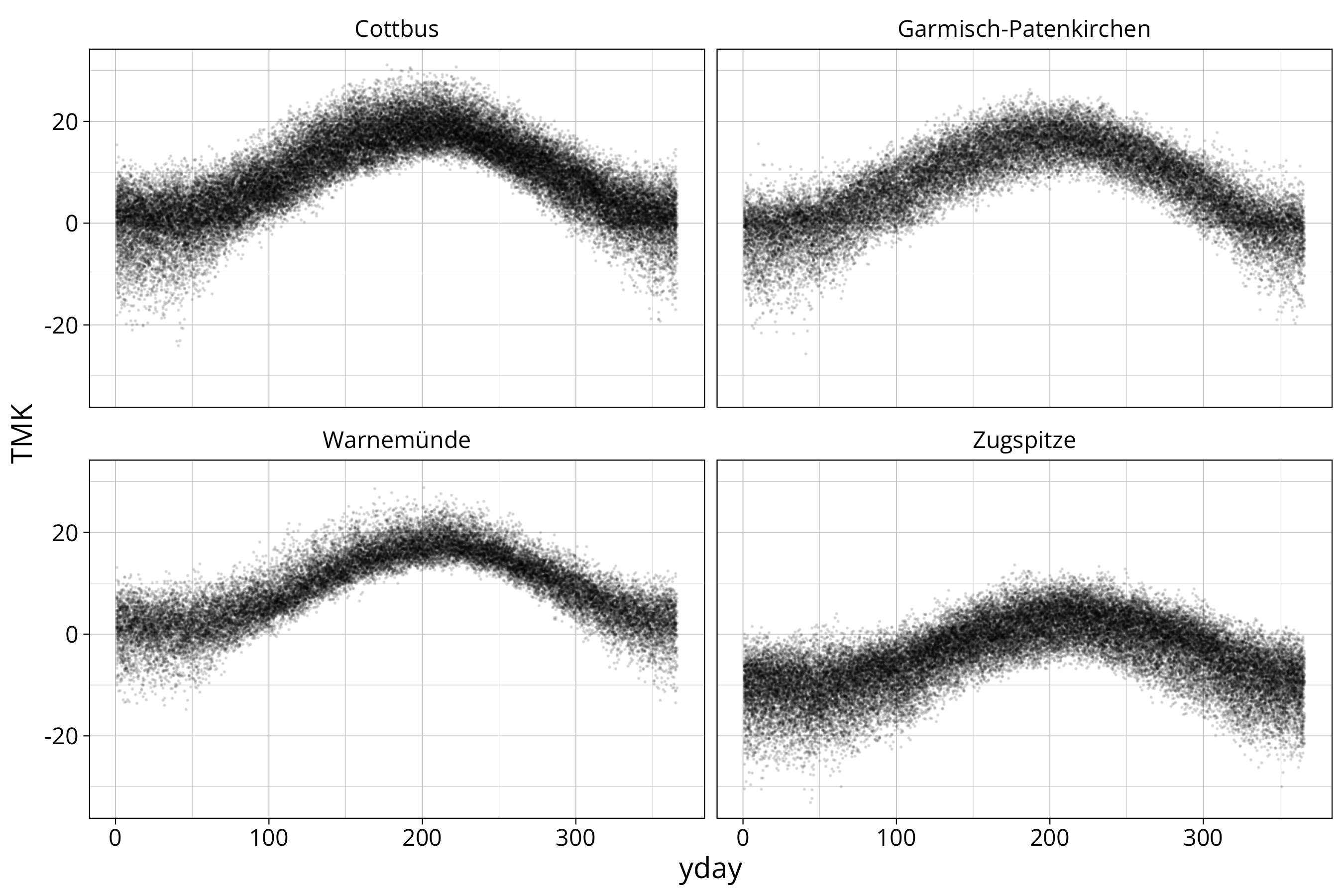
facet_wrap creates a single graph for each station.
calculate mean for month
longterm_average_month <- data %>%
group_by(STATION_NAME, month) %>%
summarise("temp longterm average" = mean(TMK, na.rm=T), "rain longterm average" = mean(RSK, na.rm=T))With group_by and summarise we again use functions from the dplyr package. The procedure is to form multiple subsets of the dataset with group_by and then calculate the summary statistics for those subsets. We group by STATION_NAME and month.
In the next plot we put all stations into one graph:
longterm_average_month %>%
ggplot() +
aes(x=month, y=`temp longterm average`, col=STATION_NAME, group=STATION_NAME) +
geom_line()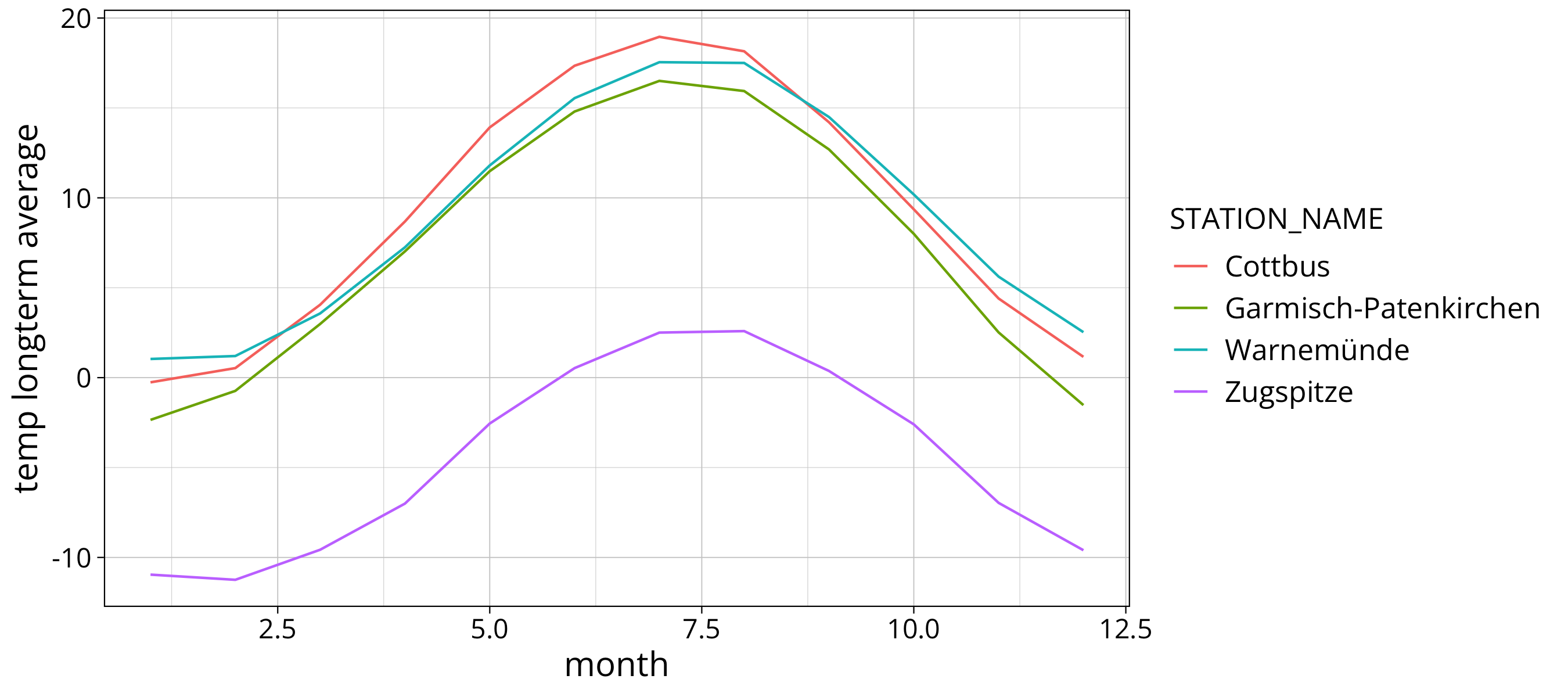
calculate mean for yday
longterm_average_yday <- data %>%
group_by(STATION_NAME, yday) %>%
summarise("temp longterm average" = mean(TMK, na.rm=T), "rain longterm average" = mean(RSK, na.rm=T))data %>%
ggplot() +
aes(x=yday, y=TMK) +
geom_point(size=0.1, alpha=0.1) +
geom_line(aes(x=yday, y=`temp longterm average`), data=longterm_average_yday, col="red") +
facet_wrap(STATION_NAME~.)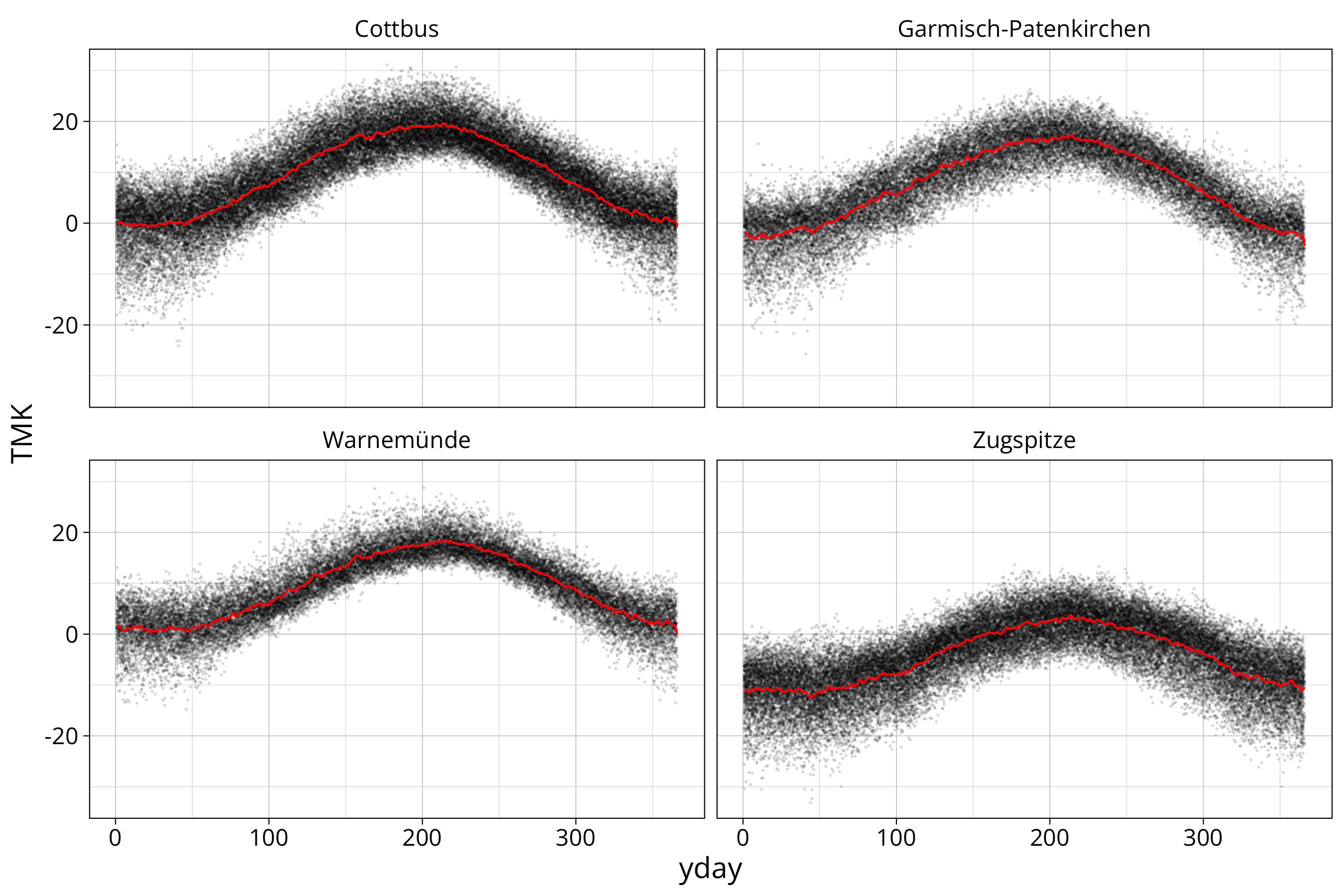
precipitation
data %>%
ggplot() +
aes(x=yday, y=RSK) +
scale_y_continuous(trans="log10") +
geom_point(size=0.1, alpha=0.1) +
facet_wrap(STATION_NAME~.)
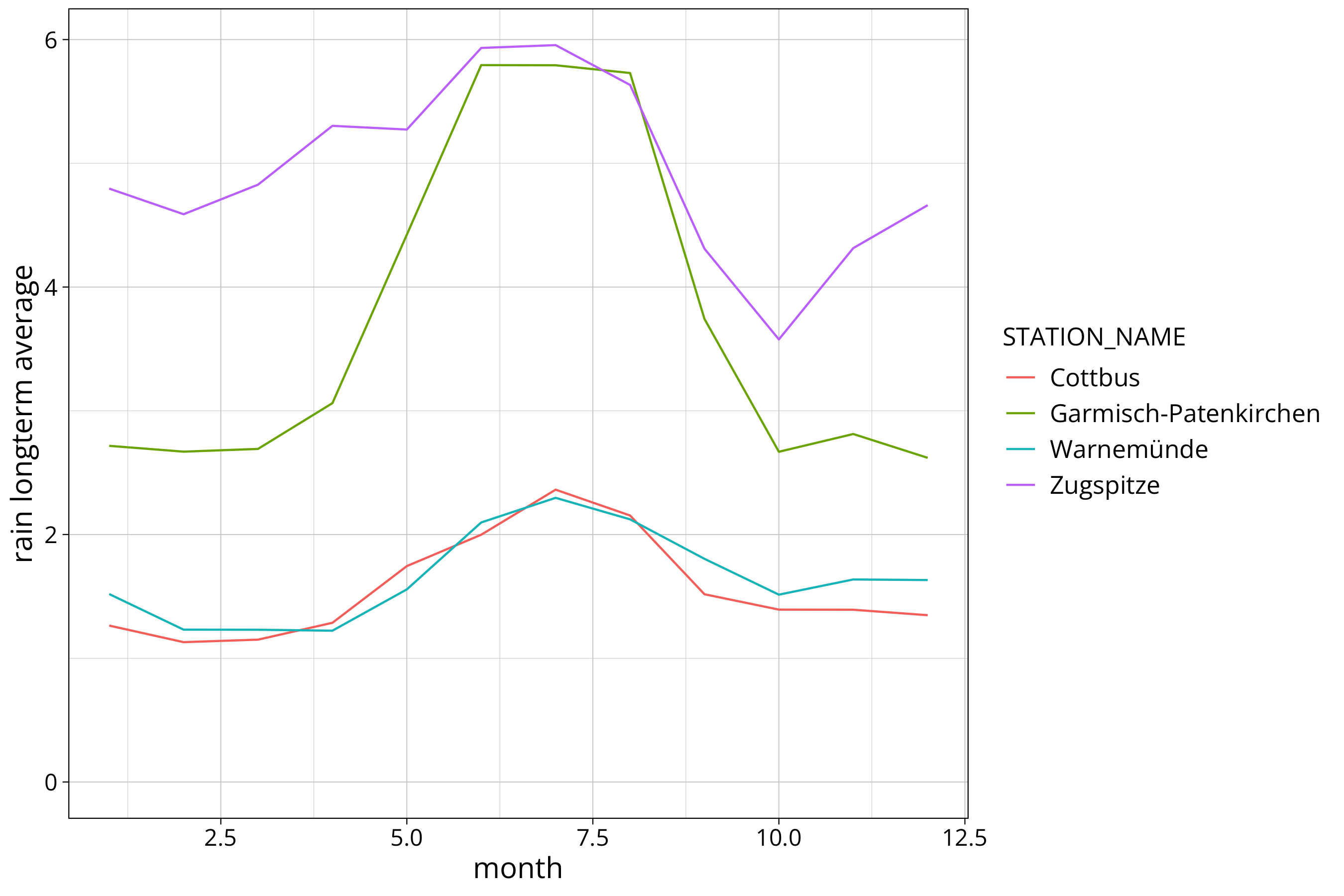
longterm_average_month %>%
ggplot() +
aes(x=month, y=`rain longterm average`, col=STATION_NAME, group=STATION_NAME) +
#scale_y_continuous(trans="log10") +
geom_line()